Talk:Region Growing Segmentation
Region Growing Segementation with Saga's Seeded Region Growing Tool UNDER CONSTRUCTION!!!!
The following tutorial explains how to delineate tree crowns, using SAGA's Seeded Region Growing Tool. The product, a polygon shapefile, can then be used in an object based classification, f.ex. in order to classify different tree species.
Material you need to complete the tutorial
Data
A multispectral image of the forest canopy A canopy height model CHM
Software
QGIS 2.18.11
SAGA 2.3.2
Split multiband image into several raster images
SAGA's Region Growing Algorithm works only with single band images. Therefor, we have to split our multiband image into its individual bands following these instructions.
Seed points
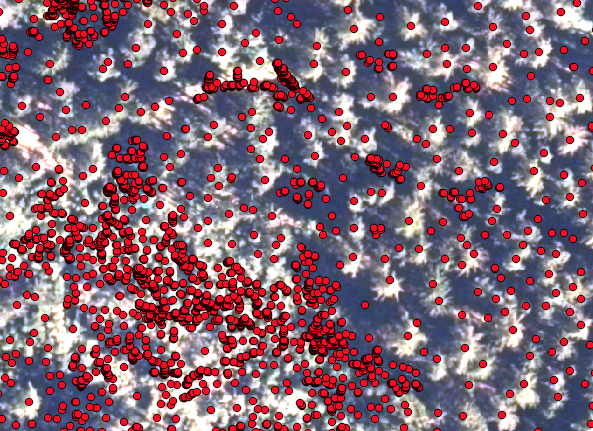
The first step here is to extract the position of the tree tops, which are going to be the starting point for the region growing algorithm. You find a description of how to derive the seed points from a CHM here . Now have a look at how the seeds align with your surface picture (orthophoto or else):
In this picture you see, that the points align well with the tree tops. However, there are many points that represent small trees (in the lower left corner) which you might not be so interested in. In order to correct that, we are going to filter the points by their height in the seed shapefile and save the shapefile with the selected seeds only (save as....)
This looks much better:
Seed point rasterization
In order to use the seed points in the region growing algorithm in SAGA, we have to convert the vector file to a raster file using the Rasterize function in QGIS. It is critical that we create a raster file that only contains the seed points and no-data. So every pixel outside the seed points has to be no-data. It is also critical that our rasterized vector image has the exact same CRS, extend and pixel size as the single band images. If this is not the case, SAGA will not be able to calculate the segments of the tree crowns.
In order to achieve this we are going to use the following settings:
Make sure that the output resolution has the exact same size as the orthophoto that you are using. Best is to copy from the properties of the raster. Choose the xtend by selecting the orthophotos extend. Nodata value should be set to 0. Next, check the properties of the newly created raster layer and load the min/max values in the styles menu.
The result should look like the universe:
Next steps:
Translate seed raster to set all pixels to no-data that are not seeds....
Next: Split stack with translate picture files
next: superimpose extend and origin
Check everything: pixels should align 100%
Next saga: growing regions algorithm
Basis of algorithm citation settings export with vectorize adjust enjoy