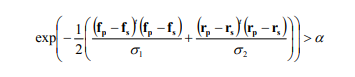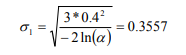Talk:Region Growing Segmentation
(→The math part) |
(→The math part) |
||
| (15 intermediate revisions by one user not shown) | |||
| Line 54: | Line 54: | ||
| − | In order to use the seed points in the region growing algorithm in SAGA, we have to convert the vector file to a raster file using the {{button|text=Rasterize}} module from the GDAL Conversions in QGIS. It is critical that we create a raster file that only contains the seed points and no-data. So every pixel outside the seed points has to be no-data. It is also critical that our rasterized vector image has the '''exact same''' CRS, extend and pixel size as the single band images. | + | In order to use the seed points in the region growing algorithm in SAGA, we have to convert the vector file to a raster file using the {{button|text=Rasterize}} module from the GDAL Conversions in QGIS. It is critical that we create a raster file that only contains the seed points and no-data. So every pixel outside the seed points has to be no-data. It is also critical that our rasterized vector image has the '''exact same''' CRS (Coordinate reference system), extend and pixel size as the single band images. |
* In the {{button|text=Processing Toolbox}}, type 'rasterize' and select the {{button|text=Rasterize}} tool ind the SAGA geoalgorithms submenu | * In the {{button|text=Processing Toolbox}}, type 'rasterize' and select the {{button|text=Rasterize}} tool ind the SAGA geoalgorithms submenu | ||
* Select the seed point shapefile as input | * Select the seed point shapefile as input | ||
| − | |||
* Select {{button|text=ID}} as {{button|text=Attribute}} and the other parameters according to the screenshot below | * Select {{button|text=ID}} as {{button|text=Attribute}} and the other parameters according to the screenshot below | ||
* Select the extend of one of the band splits as extend by clicking {{button|text=select canvas/ layers extend}} in the {{button|text=Output extend}} line | * Select the extend of one of the band splits as extend by clicking {{button|text=select canvas/ layers extend}} in the {{button|text=Output extend}} line | ||
* Copy the '''exact''' cellsize from either the metadata of one of your split images and set output raster size to {{button|text= Output resolution in map units per pixel}} | * Copy the '''exact''' cellsize from either the metadata of one of your split images and set output raster size to {{button|text= Output resolution in map units per pixel}} | ||
| − | |||
* Save to a local file | * Save to a local file | ||
| Line 83: | Line 81: | ||
| − | Now verify that the newly created seed point raster aligns 100% with one of the band split | + | Now verify that the newly created seed point raster aligns 100% with one of the band split rasters by checking the metadata or {{button|text=Save raster layer as..}} mask and by zooming into the seed pixels. Also check with the Info button[[File:Info.PNG]], if any pixel which is not a seed point, is no-data. |
| − | |||
| + | [[File:Saveas.PNG]] | ||
| − | + | Your raster should align like this: | |
| + | [[File:Seedpixel.PNG]] | ||
=Superimpose Vector= | =Superimpose Vector= | ||
| Line 197: | Line 196: | ||
=The math part= | =The math part= | ||
| − | As you see in the last image, the segments fit the crowns quite well, but there is room for improvement. You can start to fine-tune the segmentation by changing the different parameters in the segmentation tool: '''Variance in Feature Space, Variance in Position Space Similarity Treshold, Method and Neighbourhood'''. You can also try to '''include or exclude certain data'''. | + | As you see in the last image, the segments fit the crowns quite well, but there is room for improvement. You can start to fine-tune the segmentation by changing the different parameters in the segmentation tool: '''Variance in Feature Space, Variance in Position Space, (Similarity Treshold), Method and Neighbourhood'''. You can also try to '''include or exclude certain data'''. |
You can do that by trial and error, however, it helps to understand the math that lies behind these algorithms. | You can do that by trial and error, however, it helps to understand the math that lies behind these algorithms. | ||
| − | What the algorithm in the Seeded Region Growing module does, is that it starts with the seed point, goes to the adjacent pixels and joines them together if they are similar enough. 'Enough' in this case is determined by the | + | What the algorithm in the Seeded Region Growing module does, is that it starts with the seed point, goes to the adjacent pixels and joines them together if they are similar enough. 'Enough' in this case is determined by the similarity-condition which is expressed in the following formula: |
[[File:Formula_(1).PNG]] | [[File:Formula_(1).PNG]] | ||
| + | |||
| + | |||
| + | The meaning of the parameters are easy to understand: ''''f'''' is a value (f.ex. a spectral value or a height value) ''''r'''' is a location. The indices designate which value it is, the one from the seed point or from the pixel that is being investigated (should it be joined to the segment of the seed or not). | ||
| + | ''''t'''' indicates vector transposition and '''σ<sub>1</sub>''' and '''σ<sub>2</sub>''' are variances in colour and position space. The latter normalize the value differences between seed point and pixel. | ||
| + | '''α''' is the similarity criterion. It not need to be normalized and is set arbitrarily at 0.15. | ||
| + | |||
| + | '''σ<sub>1</sub>''' and '''σ<sub>2</sub>''' control how far the values (feature space / position space) are allowed to be. For a maximum distance of 40 % of the peak intensity and three color bands, the formula for '''σ<sub>1</sub>''' is: | ||
| + | |||
| + | [[File:Formula_(2).PNG]] | ||
| + | |||
| + | |||
| + | '''σ<sub>2</sub>''' determines the maximum spatial distance '''δ''' which is set to the radius of the largest crown (for spherical crowns): | ||
| + | |||
| + | [[File:Formula_(2).PNG]] | ||
| + | |||
| + | |||
| + | |||
| + | (BECHTEL 2008<ref name="bechtel">Bechtel, B. (2008)."Segmentation for Object Extraction of Trees using MATLAB and SAGA." SAGA–Seconds Out, Hamburger Beiträge Zur Physischen Geographie Und Landschaftsökologie. Univ. Hamburg, Inst. für Geographie (2008): 1-12..</ref>) | ||
| + | |||
| + | |||
| + | |||
| + | The algorithm in the Seeded Region Growing module is based on the work of ERIKSON (2003<ref name="Erikson2003">Erikson, M. (2003). Segmentation of individual tree crowns in colour aerial photographs using region growing supported by fuzzy rules. Canadian Journal of Forest Research, 33(8), 1557-1563.</ref>, 2004<ref name="Erikson2004">Erikson, M. (2004). Segmentation and classification of individual tree crowns (Vol. 320).</ref>, ERIKSON & OLOFSSON 2005<ref name="Eriolof2005">Erikson, M., & Olofsson, K. 2005). Comparison of three individual tree crown detection methods. Machine Vision and Applications, 16(4), 258-265.</ref>) | ||
| + | |||
| + | =References= | ||
| + | <references/> | ||
Latest revision as of 13:20, 10 April 2018
[edit] Region Growing Segementation with Saga's Seeded Region Growing Tool
The following tutorial explains how to delineate tree crowns, using SAGA's Seeded Region Growing Tool. The product, a polygon shapefile, can then be used in an object based classification, f.ex. in order to classify different tree species.
[edit] Material you need to complete the tutorial
Data
A multispectral image of the forest canopy
A canopy height model CHM
Software
QGIS 2.18.11
SAGA 2.3.2
[edit] Split multiband image into several raster images
SAGA's Region Growing Algorithm works only with single band images. Therefor, we have to split our multiband image into its individual bands following these instructions.
[edit] Seed points
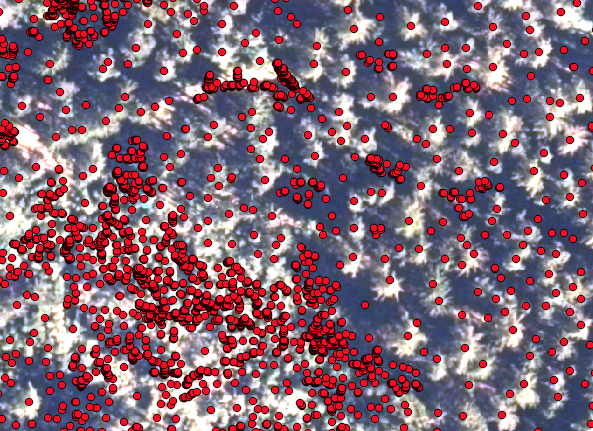
The first step here is to extract the position of the tree tops, which are going to be the starting point for the region growing algorithm. You find a description of how to derive the seed points from a CHM here . Now have a look at how the seeds align with your multiband image
In this picture you see, that the points align well with the tree tops. However, there are many points that represent small trees (in the lower left corner) which you might not be so interested in. In order to correct that, we are going to filter the points by their height in the seed shapefile and save the shapefile with the selected seeds only as a new shapefile.
If the seed points do not fit the image well, it might be due to the fact that you are using an orthophoto and not a true orthophoto. If you are working only with a smaller dataset you can help it by using the Georeferencer in the Raster menu by rubberheeting your orthophoto to make it fit. You'll find the instructions for doing so here.
This looks much better:
[edit] Seed point rasterization
In order to use the seed points in the region growing algorithm in SAGA, we have to convert the vector file to a raster file using the Rasterize module from the GDAL Conversions in QGIS. It is critical that we create a raster file that only contains the seed points and no-data. So every pixel outside the seed points has to be no-data. It is also critical that our rasterized vector image has the exact same CRS (Coordinate reference system), extend and pixel size as the single band images.
- In the Processing Toolbox, type 'rasterize' and select the Rasterize tool ind the SAGA geoalgorithms submenu
- Select the seed point shapefile as input
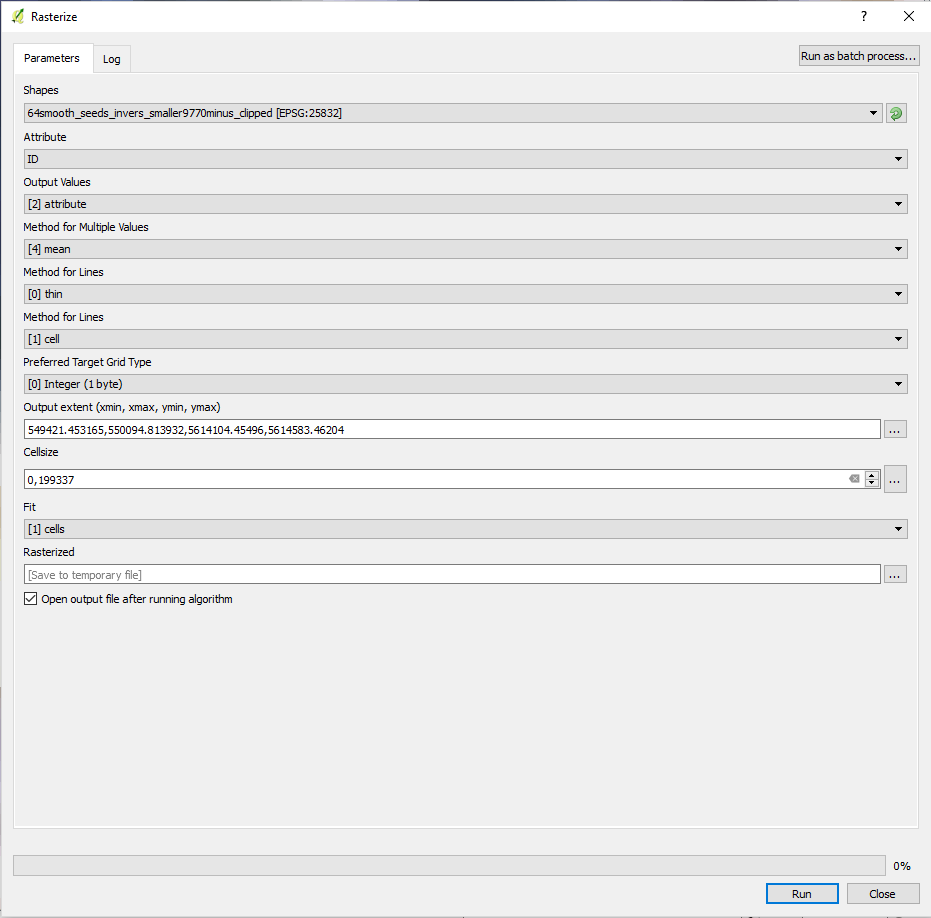
- Select ID as Attribute and the other parameters according to the screenshot below
- Select the extend of one of the band splits as extend by clicking select canvas/ layers extend in the Output extend line
- Copy the exact cellsize from either the metadata of one of your split images and set output raster size to Output resolution in map units per pixel
- Save to a local file
The result should look like this:
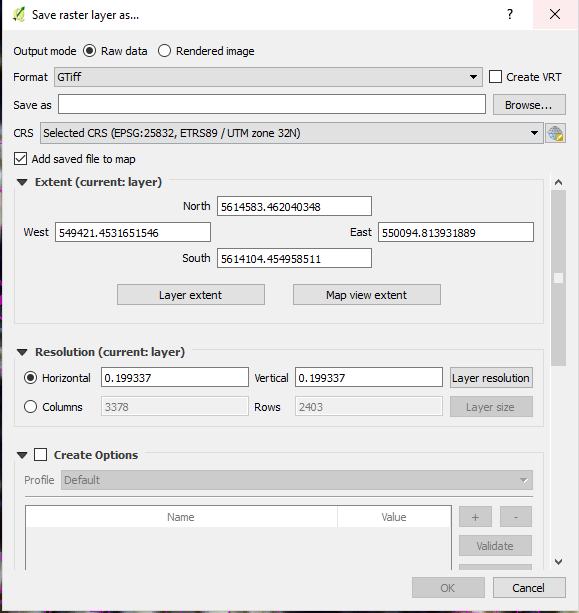
Now verify that the newly created seed point raster aligns 100% with one of the band split rasters by checking the metadata or Save raster layer as.. mask and by zooming into the seed pixels. Also check with the Info button, if any pixel which is not a seed point, is no-data.
Your raster should align like this:
[edit] Superimpose Vector
It happens regularly that, although following the previous steps meticulously, that the extend and pixel size of the rasterized vector file does not match the splitted band images 100 %. In this case we have to use the OTB Superimpose Vector tool that you can find in the Processing Toolbox.
- Open the Superimpose Vector tool in the Processing Toolbox.
- Select one of the split band images as Reference input
- Select the rasterized seed point shapefile as The image to reproject
- Fill in the other parameters as shown here:
Next, go to the Properties of the newly created layer in the Layers Panel
- Check the extend of the layer in the Metadata tab.
- Go to Styles, load max/min values and set the Color Gradient to White to black
The result should look very much the same as the first rasterized vector, but the extend and pixel size are now 100 % the same as in
the split band images.
[edit] Region Growing in SAGA
Now we finally made it to the fun part of the whole exercise, which is creating the segments around the tops of the trees i.e. the seed points. Therefor we open SAGA GIS.
- When prompted, select an empty Startup Project
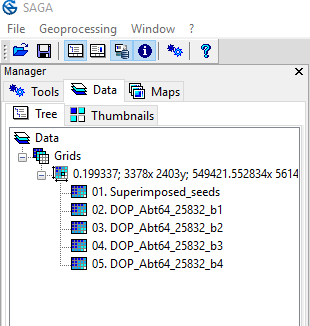
- Open all split band images and the superimposed seed raster one after another
- The layer panel on the left should now look like this:
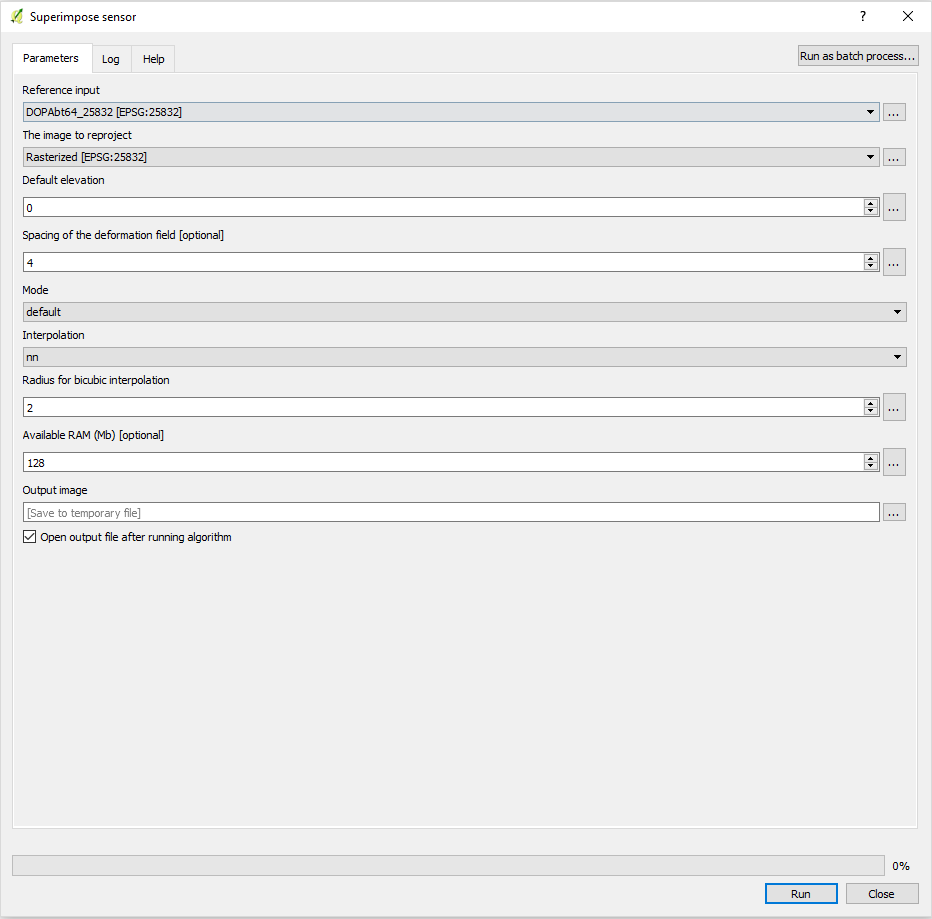
- In the line below Grids you see the extend of your raster files. If you happen to see two different of those lines, your raster images do not match 100 %. You can fix that again with the Superimpose sensor Tool in the OTB library in QGIS
[edit] Seeded Region Growing Algorithm
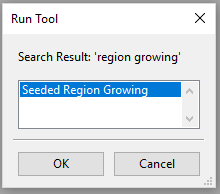
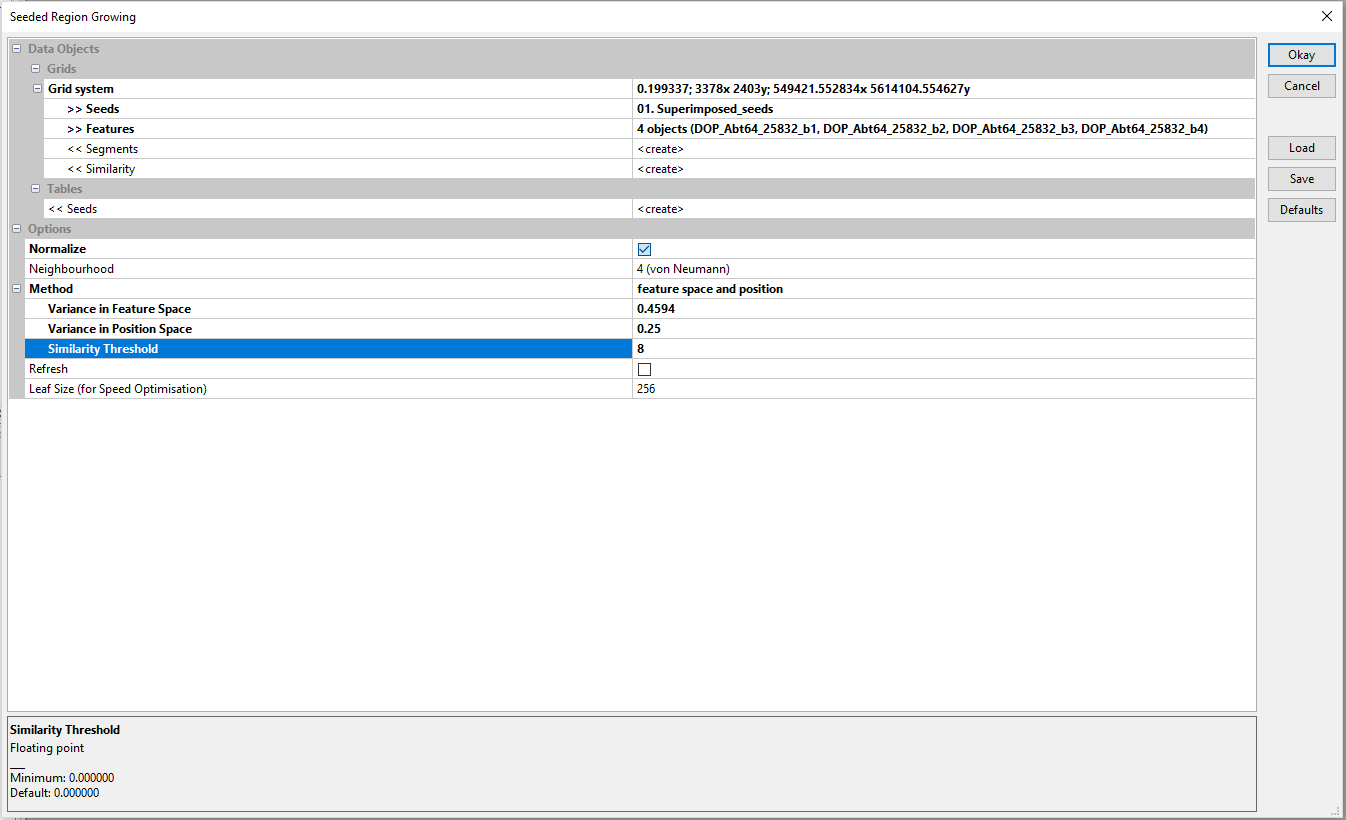
- Go to the Geoprocessing menu and select Find and Run tool
- Select the one grid on offer
- Select your seed point raster as Seeds
- Select your splitted band images as Features
- Select create in Segments, Similarity and Tables<<Seeds
- Select Normalize
- Select 4 (von Neumann
- Select feature space and position
- Insert the values shown in the picture and click Okay twice. We will talk about the input values later on...
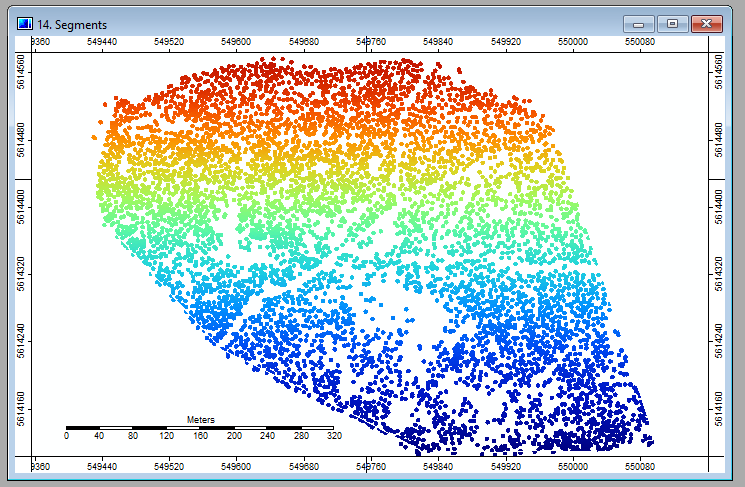
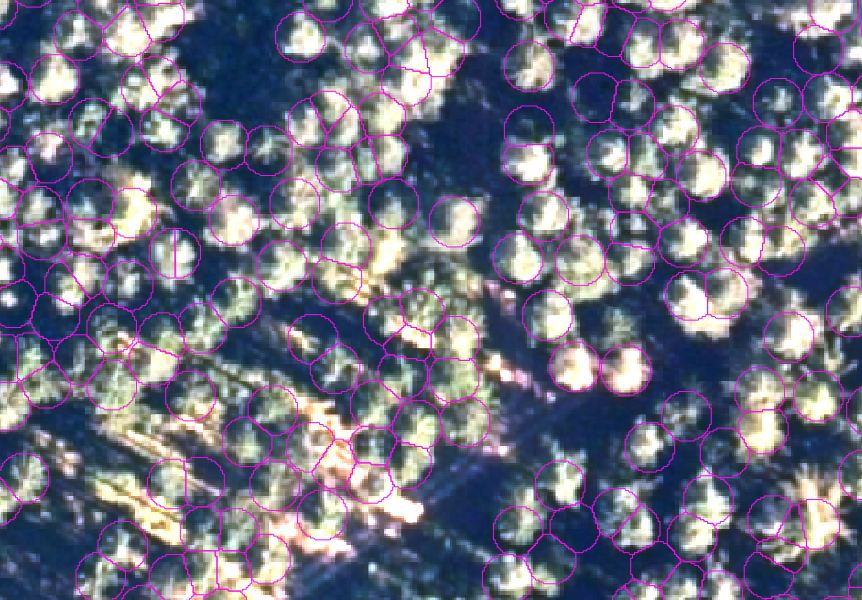
After a short while, SAGA will have finished the calculation and you'll notice two new layers in the layer panel. Double-click on Segments and you hopefully see the first fruit of all the effort that you put into this exercise:
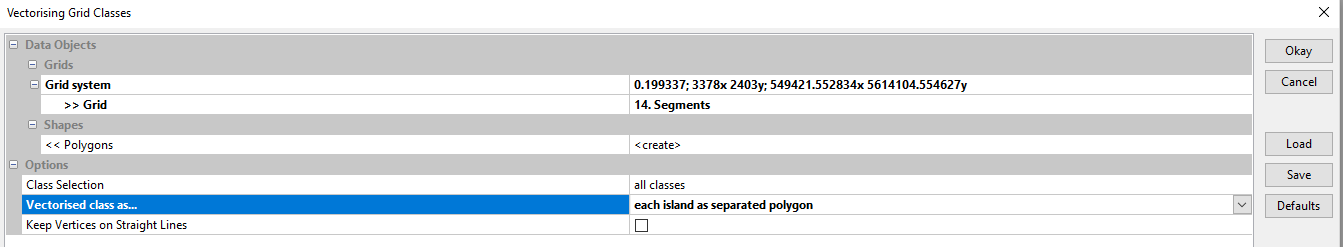
[edit] Vectorising the raster data
In the next step we are going to create polygons in a shapefile which we can then open in QGIS...
- Fill in the mask as follows and double-click Okay :
- Right click on the newly created Segments file under Shapes in the Layer panel
- Select Save as and save the file as ESRI shapefile
[edit] Opening the polygon file in QGIS
Now you can open the shapefile in QGIS and adjust the look of the segments in the Styles tab in the properties of the layer:
[edit] The math part
As you see in the last image, the segments fit the crowns quite well, but there is room for improvement. You can start to fine-tune the segmentation by changing the different parameters in the segmentation tool: Variance in Feature Space, Variance in Position Space, (Similarity Treshold), Method and Neighbourhood. You can also try to include or exclude certain data.
You can do that by trial and error, however, it helps to understand the math that lies behind these algorithms.
What the algorithm in the Seeded Region Growing module does, is that it starts with the seed point, goes to the adjacent pixels and joines them together if they are similar enough. 'Enough' in this case is determined by the similarity-condition which is expressed in the following formula:
The meaning of the parameters are easy to understand: 'f' is a value (f.ex. a spectral value or a height value) 'r' is a location. The indices designate which value it is, the one from the seed point or from the pixel that is being investigated (should it be joined to the segment of the seed or not).
't' indicates vector transposition and σ1 and σ2 are variances in colour and position space. The latter normalize the value differences between seed point and pixel.
α is the similarity criterion. It not need to be normalized and is set arbitrarily at 0.15.
σ1 and σ2 control how far the values (feature space / position space) are allowed to be. For a maximum distance of 40 % of the peak intensity and three color bands, the formula for σ1 is:
σ2 determines the maximum spatial distance δ which is set to the radius of the largest crown (for spherical crowns):
(BECHTEL 2008[1])
The algorithm in the Seeded Region Growing module is based on the work of ERIKSON (2003[2], 2004[3], ERIKSON & OLOFSSON 2005[4])
[edit] References
- ↑ Bechtel, B. (2008)."Segmentation for Object Extraction of Trees using MATLAB and SAGA." SAGA–Seconds Out, Hamburger Beiträge Zur Physischen Geographie Und Landschaftsökologie. Univ. Hamburg, Inst. für Geographie (2008): 1-12..
- ↑ Erikson, M. (2003). Segmentation of individual tree crowns in colour aerial photographs using region growing supported by fuzzy rules. Canadian Journal of Forest Research, 33(8), 1557-1563.
- ↑ Erikson, M. (2004). Segmentation and classification of individual tree crowns (Vol. 320).
- ↑ Erikson, M., & Olofsson, K. 2005). Comparison of three individual tree crown detection methods. Machine Vision and Applications, 16(4), 258-265.